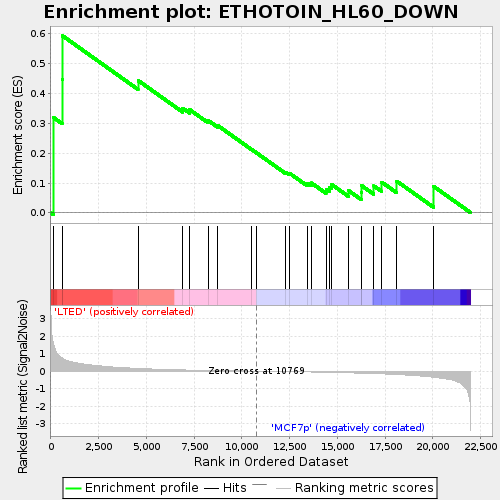
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GE_agg.GE_agg.cls#LTED_versus_MCF7p.GE_agg.cls#LTED_versus_MCF7p_repos |
| Phenotype | GE_agg.cls#LTED_versus_MCF7p_repos |
| Upregulated in class | LTED |
| GeneSet | ETHOTOIN_HL60_DOWN |
| Enrichment Score (ES) | 0.5935287 |
| Normalized Enrichment Score (NES) | 1.4849267 |
| Nominal p-value | 0.056603774 |
| FDR q-value | 0.70415634 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | 586 | NA | BCAT1 Entrez, Source | branched chain amino-acid transaminase 1, cytosolic | 120 | 1.628 | 0.3203 | Yes |
| 2 | 1130 | NA | LYST Entrez, Source | lysosomal trafficking regulator | 586 | 0.748 | 0.4489 | Yes |
| 3 | 4242 | NA | MFNG Entrez, Source | MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | 613 | 0.729 | 0.5935 | Yes |
| 4 | 23039 | NA | XPO7 Entrez, Source | exportin 7 | 4563 | 0.151 | 0.4441 | No |
| 5 | 9057 | NA | SLC7A6 Entrez, Source | solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 | 6896 | 0.067 | 0.3515 | No |
| 6 | 1660 | NA | DHX9 Entrez, Source | DEAH (Asp-Glu-Ala-His) box helicase 9 | 7269 | 0.057 | 0.3460 | No |
| 7 | 4144 | NA | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 8227 | 0.039 | 0.3103 | No |
| 8 | 8795 | NA | TNFRSF10B Entrez, Source | tumor necrosis factor receptor superfamily, member 10b | 8744 | 0.029 | 0.2927 | No |
| 9 | 1629 | NA | DBT Entrez, Source | dihydrolipoamide branched chain transacylase E2 | 10520 | 0.003 | 0.2125 | No |
| 10 | 2799 | NA | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase | 10758 | 0.000 | 0.2018 | No |
| 11 | 5273 | NA | SERPINB10 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 10 | 12286 | -0.024 | 0.1370 | No |
| 12 | 26121 | NA | PRPF31 Entrez, Source | pre-mRNA processing factor 31 | 12492 | -0.027 | 0.1332 | No |
| 13 | 4170 | NA | MCL1 Entrez, Source | myeloid cell leukemia 1 | 13429 | -0.046 | 0.0999 | No |
| 14 | 1314 | NA | COPA Entrez, Source | coatomer protein complex, subunit alpha | 13640 | -0.051 | 0.1006 | No |
| 15 | 4751 | NA | NEK2 Entrez, Source | NIMA-related kinase 2 | 14408 | -0.068 | 0.0793 | No |
| 16 | 1654 | NA | DDX3X Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked | 14574 | -0.072 | 0.0862 | No |
| 17 | 6120 | NA | RPE Entrez, Source | ribulose-5-phosphate-3-epimerase | 14679 | -0.075 | 0.0964 | No |
| 18 | 3308 | NA | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 15581 | -0.098 | 0.0751 | No |
| 19 | 8295 | NA | TRRAP Entrez, Source | transformation/transcription domain-associated protein | 16251 | -0.119 | 0.0684 | No |
| 20 | 10672 | NA | GNA13 Entrez, Source | guanine nucleotide binding protein (G protein), alpha 13 | 16266 | -0.119 | 0.0917 | No |
| 21 | 699 | NA | BUB1 Entrez, Source | BUB1 mitotic checkpoint serine/threonine kinase | 16913 | -0.141 | 0.0905 | No |
| 22 | 835 | NA | CASP2 Entrez, Source | caspase 2, apoptosis-related cysteine peptidase | 17325 | -0.157 | 0.1033 | No |
| 23 | 10613 | NA | ERLIN1 Entrez, Source | ER lipid raft associated 1 | 18114 | -0.195 | 0.1064 | No |
| 24 | 10548 | NA | TM9SF1 Entrez, Source | transmembrane 9 superfamily member 1 | 20046 | -0.351 | 0.0889 | No |