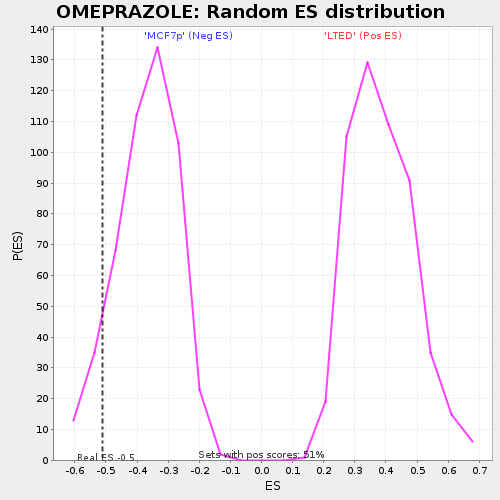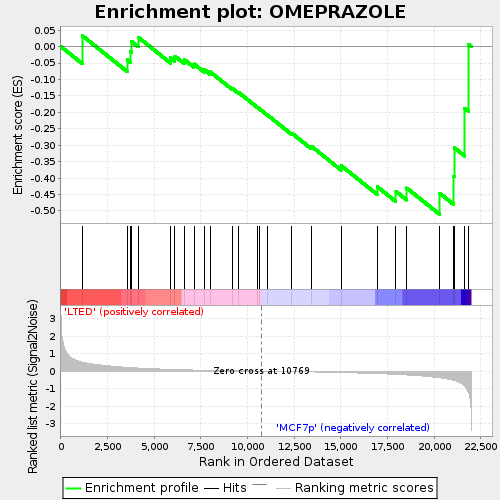
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GE_agg.GE_agg.cls#LTED_versus_MCF7p.GE_agg.cls#LTED_versus_MCF7p_repos |
| Phenotype | GE_agg.cls#LTED_versus_MCF7p_repos |
| Upregulated in class | MCF7p |
| GeneSet | OMEPRAZOLE |
| Enrichment Score (ES) | -0.5108652 |
| Normalized Enrichment Score (NES) | -1.3893152 |
| Nominal p-value | 0.08571429 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | 1544 | NA | CYP1A2 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 2 | 1159 | 0.511 | 0.0325 | No |
| 2 | 4780 | NA | NFE2L2 Entrez, Source | nuclear factor, erythroid 2-like 2 | 3546 | 0.213 | -0.0406 | No |
| 3 | 7421 | NA | VDR Entrez, Source | vitamin D (1,25- dihydroxyvitamin D3) receptor | 3726 | 0.201 | -0.0151 | No |
| 4 | 4351 | NA | MPI Entrez, Source | mannose phosphate isomerase | 3769 | 0.198 | 0.0160 | No |
| 5 | 367 | NA | AR Entrez, Source | androgen receptor | 4147 | 0.175 | 0.0281 | No |
| 6 | 9682 | NA | KDM4A Entrez, Source | lysine (K)-specific demethylase 4A | 5878 | 0.099 | -0.0341 | No |
| 7 | 496 | NA | ATP4B Entrez, Source | ATPase, H+/K+ exchanging, beta polypeptide | 6103 | 0.091 | -0.0291 | No |
| 8 | 10919 | NA | EHMT2 Entrez, Source | euchromatic histone-lysine N-methyltransferase 2 | 6620 | 0.076 | -0.0399 | No |
| 9 | 10951 | NA | CBX1 Entrez, Source | chromobox homolog 1 | 7130 | 0.061 | -0.0530 | No |
| 10 | 4137 | NA | MAPT Entrez, Source | microtubule-associated protein tau | 7674 | 0.049 | -0.0695 | No |
| 11 | 196 | NA | AHR Entrez, Source | aryl hydrocarbon receptor | 8000 | 0.043 | -0.0771 | No |
| 12 | 1565 | NA | CYP2D6 Entrez, Source | cytochrome P450, family 2, subfamily D, polypeptide 6 | 9172 | 0.023 | -0.1266 | No |
| 13 | 1559 | NA | CYP2C9 Entrez, Source | cytochrome P450, family 2, subfamily C, polypeptide 9 | 9513 | 0.017 | -0.1392 | No |
| 14 | 5347 | NA | PLK1 Entrez, Source | polo-like kinase 1 | 10514 | 0.003 | -0.1841 | No |
| 15 | 7201 | NA | TRHR Entrez, Source | thyrotropin-releasing hormone receptor | 10622 | 0.002 | -0.1887 | No |
| 16 | 1557 | NA | CYP2C19 Entrez, Source | cytochrome P450, family 2, subfamily C, polypeptide 19 | 11076 | -0.004 | -0.2086 | No |
| 17 | 495 | NA | ATP4A Entrez, Source | ATPase, H+/K+ exchanging, alpha polypeptide | 12378 | -0.025 | -0.2636 | No |
| 18 | 4170 | NA | MCL1 Entrez, Source | myeloid cell leukemia 1 | 13429 | -0.046 | -0.3036 | No |
| 19 | 2671 | NA | GFER Entrez, Source | growth factor, augmenter of liver regeneration | 15010 | -0.083 | -0.3617 | No |
| 20 | 29994 | NA | BAZ2B Entrez, Source | bromodomain adjacent to zinc finger domain, 2B | 16946 | -0.142 | -0.4260 | No |
| 21 | 3315 | NA | HSPB1 Entrez, Source | heat shock 27kDa protein 1 | 17953 | -0.187 | -0.4405 | No |
| 22 | 1132 | NA | CHRM4 Entrez, Source | cholinergic receptor, muscarinic 4 | 18528 | -0.219 | -0.4300 | No |
| 23 | 1576 | NA | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 20305 | -0.385 | -0.4466 | Yes |
| 24 | 2908 | NA | NR3C1 Entrez, Source | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 21034 | -0.516 | -0.3936 | Yes |
| 25 | 3248 | NA | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 21086 | -0.528 | -0.3078 | Yes |
| 26 | 5243 | NA | ABCB1 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 21640 | -0.868 | -0.1880 | Yes |
| 27 | 5468 | NA | PPARG Entrez, Source | peroxisome proliferator-activated receptor gamma | 21841 | -1.224 | 0.0072 | Yes |