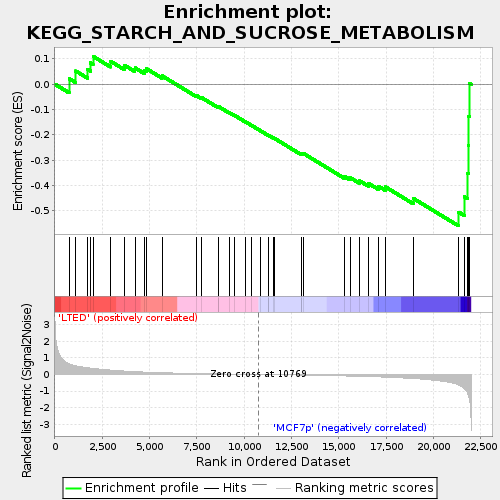
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GE_agg.GE_agg.cls#LTED_versus_MCF7p.GE_agg.cls#LTED_versus_MCF7p_repos |
| Phenotype | GE_agg.cls#LTED_versus_MCF7p_repos |
| Upregulated in class | MCF7p |
| GeneSet | KEGG_STARCH_AND_SUCROSE_METABOLISM |
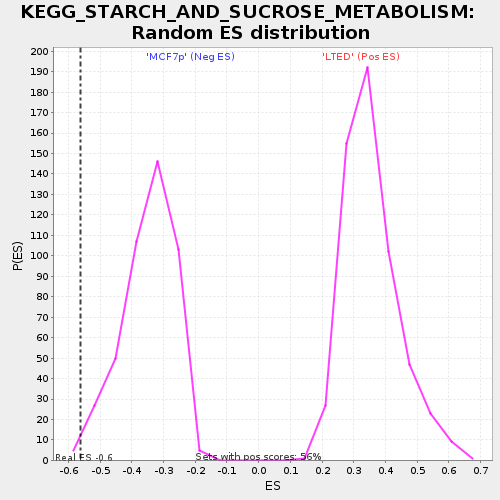
| Enrichment Score (ES) | -0.5605355 |
| Normalized Enrichment Score (NES) | -1.6032592 |
| Nominal p-value | 0.0112866815 |
| FDR q-value | 0.12308301 |
| FWER p-Value | 0.509 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | 278 | NA | AMY1C Entrez, Source | amylase, alpha 1C (salivary) | 749 | 0.652 | 0.0224 | No |
| 2 | 283209 | NA | PGM2L1 Entrez, Source | phosphoglucomutase 2-like 1 | 1096 | 0.528 | 0.0524 | No |
| 3 | 2538 | NA | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 1729 | 0.402 | 0.0585 | No |
| 4 | 5167 | NA | ENPP1 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 1884 | 0.380 | 0.0845 | No |
| 5 | 57818 | NA | G6PC2 Entrez, Source | glucose-6-phosphatase, catalytic, 2 | 2016 | 0.365 | 0.1101 | No |
| 6 | 7364 | NA | UGT2B7 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide B7 | 2941 | 0.261 | 0.0907 | No |
| 7 | 2632 | NA | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 | 3685 | 0.204 | 0.0746 | No |
| 8 | 2997 | NA | GYS1 Entrez, Source | glycogen synthase 1 (muscle) | 4230 | 0.170 | 0.0646 | No |
| 9 | 3099 | NA | HK2 Entrez, Source | hexokinase 2 | 4730 | 0.144 | 0.0543 | No |
| 10 | 5236 | NA | PGM1 Entrez, Source | phosphoglucomutase 1 | 4846 | 0.139 | 0.0611 | No |
| 11 | 55276 | NA | PGM2 Entrez, Source | phosphoglucomutase 2 | 5661 | 0.106 | 0.0333 | No |
| 12 | 3098 | NA | HK1 Entrez, Source | hexokinase 1 | 7481 | 0.053 | -0.0450 | No |
| 13 | 6476 | NA | SI Entrez, Source | sucrase-isomaltase (alpha-glucosidase) | 7720 | 0.048 | -0.0516 | No |
| 14 | 57733 | NA | GBA3 Entrez, Source | glucosidase, beta, acid 3 (gene/pseudogene) | 8617 | 0.031 | -0.0897 | No |
| 15 | 2548 | NA | GAA Entrez, Source | glucosidase, alpha; acid | 8624 | 0.031 | -0.0873 | No |
| 16 | 5169 | NA | ENPP3 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 3 | 9212 | 0.022 | -0.1121 | No |
| 17 | 5836 | NA | PYGL Entrez, Source | phosphorylase, glycogen, liver | 9503 | 0.018 | -0.1238 | No |
| 18 | 11181 | NA | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 10055 | 0.010 | -0.1480 | No |
| 19 | 7358 | NA | UGDH Entrez, Source | UDP-glucose 6-dehydrogenase | 10380 | 0.005 | -0.1623 | No |
| 20 | 7363 | NA | UGT2B4 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide B4 | 10861 | -0.001 | -0.1841 | No |
| 21 | 2998 | NA | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 11287 | -0.008 | -0.2028 | No |
| 22 | 2645 | NA | GCK Entrez, Source | glucokinase (hexokinase 4) | 11308 | -0.008 | -0.2030 | No |
| 23 | 10941 | NA | UGT2A1 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus | 11530 | -0.011 | -0.2121 | No |
| 24 | 79799 | NA | UGT2A3 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide A3 | 11602 | -0.012 | -0.2143 | No |
| 25 | 80146 | NA | UXS1 Entrez, Source | UDP-glucuronate decarboxylase 1 | 13025 | -0.038 | -0.2757 | No |
| 26 | 5837 | NA | PYGM Entrez, Source | phosphorylase, glycogen, muscle | 13048 | -0.038 | -0.2734 | No |
| 27 | 178 | NA | AGL Entrez, Source | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase | 13141 | -0.040 | -0.2741 | No |
| 28 | 2821 | NA | GPI Entrez, Source | glucose-6-phosphate isomerase | 15322 | -0.091 | -0.3655 | No |
| 29 | 8972 | NA | MGAM Entrez, Source | maltase-glucoamylase (alpha-glucosidase) | 15608 | -0.099 | -0.3698 | No |
| 30 | 5834 | NA | PYGB Entrez, Source | phosphorylase, glycogen; brain | 16102 | -0.114 | -0.3823 | No |
| 31 | 7360 | NA | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 16570 | -0.129 | -0.3924 | No |
| 32 | 54490 | NA | UGT2B28 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide B28 | 17103 | -0.148 | -0.4038 | No |
| 33 | 2990 | NA | GUSB Entrez, Source | glucuronidase, beta | 17468 | -0.164 | -0.4062 | No |
| 34 | 3101 | NA | HK3 Entrez, Source | hexokinase 3 (white cell) | 18938 | -0.247 | -0.4516 | No |
| 35 | 2595 | NA | GANC Entrez, Source | glucosidase, alpha; neutral C | 21331 | -0.633 | -0.5056 | Yes |
| 36 | 7365 | NA | UGT2B10 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide B10 | 21635 | -0.862 | -0.4447 | Yes |
| 37 | 54578 | NA | UGT1A6 Entrez, Source | UDP glucuronosyltransferase 1 family, polypeptide A6 | 21819 | -1.162 | -0.3522 | Yes |
| 38 | 10720 | NA | UGT2B11 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide B11 | 21855 | -1.284 | -0.2424 | Yes |
| 39 | 7366 | NA | UGT2B15 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide B15 | 21874 | -1.338 | -0.1272 | Yes |
| 40 | 54576 | NA | UGT1A8 Entrez, Source | UDP glucuronosyltransferase 1 family, polypeptide A8 | 21915 | -1.531 | 0.0038 | Yes |